The department of Biological Sciences at Rensselaer encompasses a diverse and interdisciplinary portfolio of research. Our faculty, graduate students and undergraduates are exploring areas at the forefront of modern biology and biophysics. A list of all active research groups currently accepting Ph.D. graduate students can be found below.
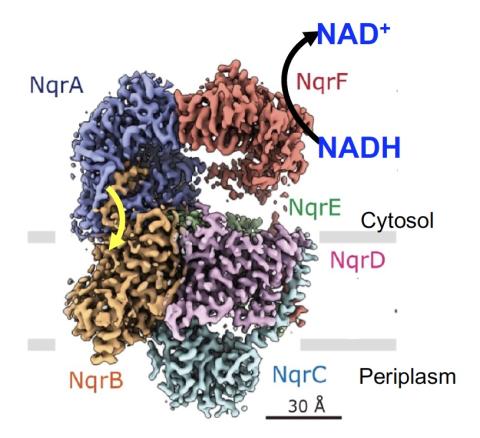
Barquera Lab
PI: Blanca Barquera
The Barquera lab studies membrane proteins responsible for energy metabolism in beneficial and pathogenic bacteria.
Key words: Bacteria, energy metabolism, membrane proteins, gut bacteria, pathogenic bacteria, ion gradients, respiration
Lab Location: CBIS 2444
barqub@rpi.edu
Google Scholar
Faculty Bio Page
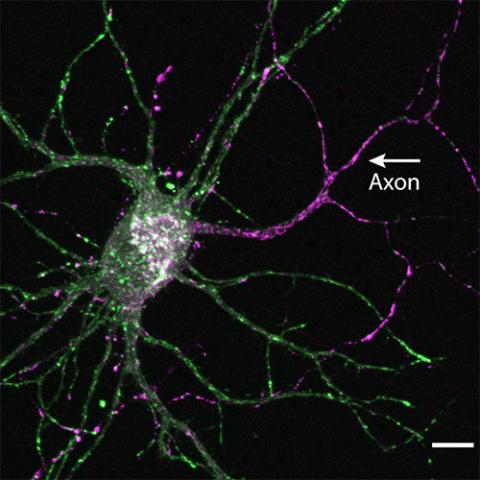
Bentley Lab
PI: Marvin Bentley
The Bentley lab studies the mechanisms of intracellular trafficking that maintain neurons.
Key words: Cell biology, neurons, vesicle, cytoskeleton, transport, kinesin, polarity, cell biology, live-cell microscopy, tissue culture, molecular biology.
Lab Location: CBIS 2332
Google Scholar
Faculty Bio Page
Bystroff Lab
PI: Chris Bystroff
The Bystroff lab is developing fluorescent biosensors and a contraceptive vaccine, using computational protein design.
Key words: immunocontraception, protein design, folding, assembly, fluorescent proteins.
Lab Location: JROWL 3C07
Lab website
Google Scholar
Faculty Bio Page
 Forth Lab
Forth Lab
PI: Scott Forth
The Forth lab uses biophysical tools to understand how cytoskeletal networks generate forces during mitosis and neuronal development.
Key words: biophysics, mechanics, force, microtubules, MAPs, kinesins, mitosis, neurons, tau, optical trapping, neurons
Lab Location: CBIS 2444
Lab website
Google Scholar
Faculty Bio Page
 Hurley Lab
Hurley Lab
PI: Jennifer Marie Hurley
The Hurley lab's research focus is on the fundamental mechanisms underlying circadian rhythms and the functions these rhythms control on the metabolic and immunological levels.
Key words: Circadian, Clock, Neurospora crassa, Mus musculus, immunology, metabolism, biofuels, Alzheimer's Disease, disordered proteins, macrophages
Lab Location: CBIS 2440
Lab website
Google Scholar
Faculty Bio Page
Larson Lab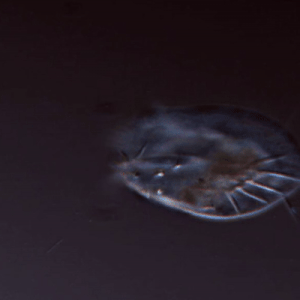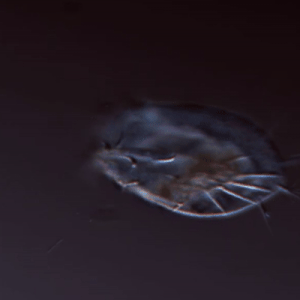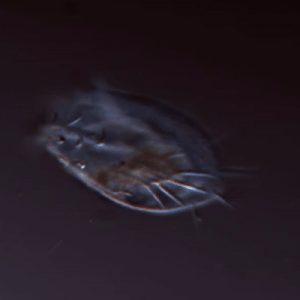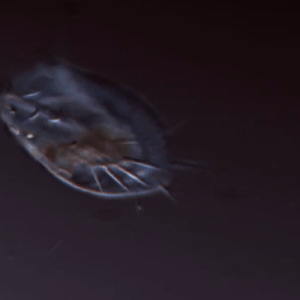
PI: Ben Larson
The Larson lab’s research focuses on the regulation and evolution of complex cellular function in microbial eukaryotes.
Keywords: cell shape, cell movement, protists, microscopy, machine learning, complex cellular behavior, evolutionary biology
Lab location: CBIS 2332
Lab website
Google Scholar
Faculty Bio Page
Makhatadze Lab
PI: George Makhatadze
The Makhatadze Lab uses biophysical methods and modern computational tools to study (1) protein folding, misfolding and amyloidogenesis and (2) evolutionary adaptation of proteins to extreme environmental conditions such as temperature, pH, salt, and hydrostatic pressure.
Key words: Protein Folding and Stability; Protein Misfolding and Amyloidogenesis; Functional Protein Dynamics; Protein Evolution; Mechanisms of Adaptation to Extreme Temperature and Pressure; Biophysical Methods; Computational Methods; Molecular Dynamics Simulations
Lab Location: CBIS 3244A
Google Scholar
Faculty Bio Page

Nierzwicki-Bauer Lab
PI: Sandra Nierzwicki-Bauer
Research in the Nierzwicki-Bauer lab is primarily focused on aquatic biota, from microbes to fish, as well as the abiotic and biotic factors that impact them and the ecosystem.
Key words: Aquatic biota, Azolla-Anabaena symbiosis, Freshwater security, In-situ remote sensing, Invasive species, Microbial communities
Lab Locations: Material Research Center and Darrin Fresh Water Institute
Google Scholar
Faculty Bio Page

Rose Lab
PI: Kevin Rose
The Rose lab's interdisciplinary research draws on diverse skills in biology, ecology, biogeochemistry, advanced environmental sensors, and computational modeling.
Key words: aquatic ecology, water, ecosystems, global change, lakes
Lab Location: JRSC 3C17
Lab website
Google Scholar
Faculty Bio Page
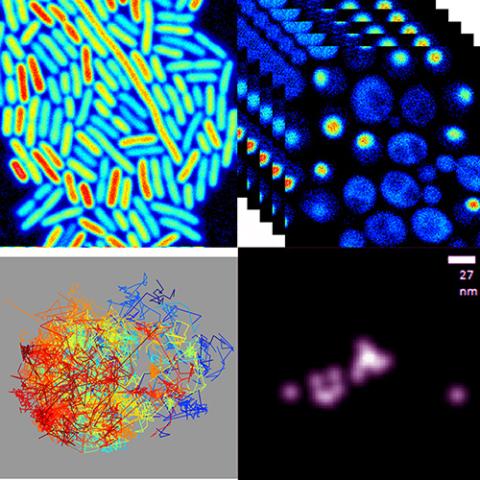
Royer Lab
PI: Catherine A. Royer
The Royer lab uses experimental and computational biophysical tools to probe the mechanisms that dictate biomolecular conformational landscapes and cell state transitions.
Key words: protein dynamics, protein conformational landscapes, cell size control, high pressure, extremophiles, fluorescence, NMR, Small angle x-ray scattering, molecular dynamics simulations
Lab Location: CBIS 3244B
Lab website
Google Scholar
Faculty Bio Page
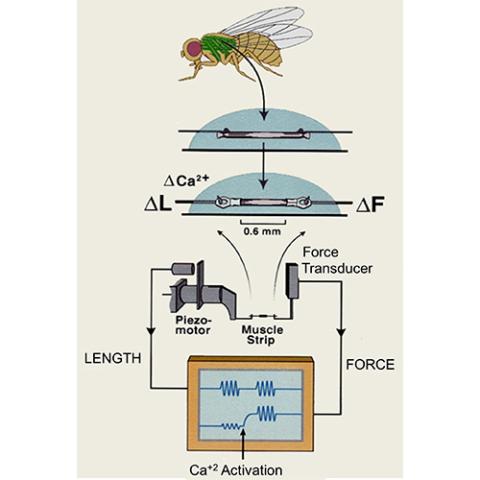
Swank Lab
PI: Douglas Swank
The Swank lab investigates muscle diseases, development, aging and mechanical properties such as stretch activation.
Key words: Drosophila, muscle, muscle mechanics, stretch activation, sarcopenia, hypertrophic cardiomyopathy, aging, muscle stem cells, myosin
Lab Location: CBIS 2332
Lab website
Google Scholar
Faculty Bio Page
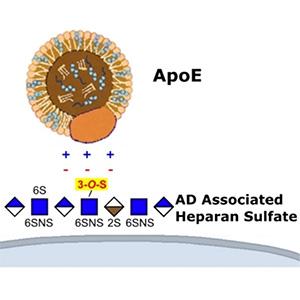
Wang Lab
PI: Chunyu Wang
Research within the Wang Lab focuses on the structural mechanisms and drug discovery in Alzheimer’s disease (AD), Hedgehog signaling and p53 in cancer, which have tremendous impact on human health.
Key words: Alzheimer's disease, NMR, tau, heparan sulfate, ApoE, amyloid, hedgehog, p53
Lab Location: CBIS 4332
Lab website
Google Scholar
Faculty Bio Page
